Introduction. The recent Ebola outbreak in West Africa has now reached historic proportions and underscores the vulnerability of populations worldwide to pathogens for which no vaccine or therapeutic agent exists. More than 1,400 deaths and 2,600 cases have been reported in four neighboring countries as of August, 22, 2014 by WHO. Ebola virus disease (EVD), also known as Ebola hemorrhagic fever (EHF), is caused by five known species within the genus Ebolavirus, in the Filovirus family. Outbreaks of EVD have occurred in Africa in the past [http://www.cdc.gov/vhf/ebola/resources/outbreak-table.html], however the current epidemic caused by Zaire Ebolavirus [Baize et al., 2014], has been characterized by greater breadth and rapid spread (reported cases and geographically). Not surprisingly, there is renewed urgency for research related to therapeutic agent and vaccine development. Herein we present a summary of the major findings for EBOV-related epitope data captured to date in the IEDB.
Summary of T and B cell epitopes IEDB data by Filoviridae family species. As of August 10, 2014, the IEDB reported 67 T cell (CD4+ and CD8+) and 35 B cell epitopes (linear and conformational), from viruses within the Ebolavirus and Marburgvirus genera. Within the genus Ebolavirus (EBOV), data are provided for all known species, including Zaire, Sudan, Reston, Bundibugyo and Tai Forest Ebolavirus, as well as two unclassified species (Ebola virus Yambio 0403 and Ebola virus sp.). Of the 142 total Filovirus-related B and T cell epitopes, the EBOV epitopes represent 71% (101 epitopes), with Zaire Ebolavirus being the most represented [Table 1]. To date, 29 papers have been published that describe experimental data on the epitopes in the Filoviridae family, with 23 papers focused on the EBOV-related epitope data.

EBOV-related epitope data. The EBOV genome encodes seven proteins: envelope glycoprotein (GP), nucleoprotein (NP), polymerase cofactor (VP35), transcription activator (VP30), matrix protein (VP40), secondary matrix protein (VP24), and RNA polymerase (L). The greatest percentage of epitopes has been defined for the GP (56; 54%) and NP (34; 32%) proteins; however, all proteins except the L protein are represented in the epitope data [Figure 1].

Figure 2 shows the breakdown of data by protein and response type (B cell or T cell and CD4 vs. CD8). While B and T cell epitopes have been defined for GP and NP, far fewer T cell responses are reported for the other four EBOV proteins (VP24, VP30, VP35 and VP40), with a complete absence of B cell data. There are currently 44 T cell and 15 B cell epitopes for GP and 10 T cell and 20 B cell epitopes for NP. Note, not all T cell epitopes are fully defined at the level of CD4/CD8 for GP, NP, VP30 and VP35 as shown in Figure 2 inset. Negative data are also available in the IEDB (data not shown).

Unexpectedly, human and non-human primate hosts are absent in the T and B-cell assays. Here we see that murine hosts predominate and a smaller number of rabbit studies have been reported only for antibodies.
Of the monoclonal antibodies (mAbs) described to date, only one was derived from a human survivor of EHF of the 1995 Kikwit outbreak (mAb KZ52) [Table 2]. Analysis of the GP protein epitopes identified using assays demonstrating in vitro correlates of protection (virus neutralization) or in vivo survival assays, so-called ‘functional epitopes,’ revealed seven mAbs, including mAb 4G7 (ZMab), which is present in the recently administered treatment cocktail, ZMapp [Mapp Biopharmaceuticals]. Two of these epitopes were identified from also the X-ray structures of the GP trimer in complexes with mAb 16F6, the first antibody known to neutralize Sudan virus, [PDB: 3VE0] and KZ52 [PDB: 3CSY]. For the NP protein, the only epitope was identified from the virus neutralizing assay using a polyclonal antibody [Table 2].
Three B cell epitopes were reported for the GP1 protein of Marburgvirus: two linear epitopes with IEDB IDs of 7674 and 150883, were identified for the mAbs 3H5 and 7G8, respectively, using a protection assay via passive transfer, and one linear epitope, 181232, for the mAbs APH159-1-3 and MGP72-17, using a virus neutralizing assay.
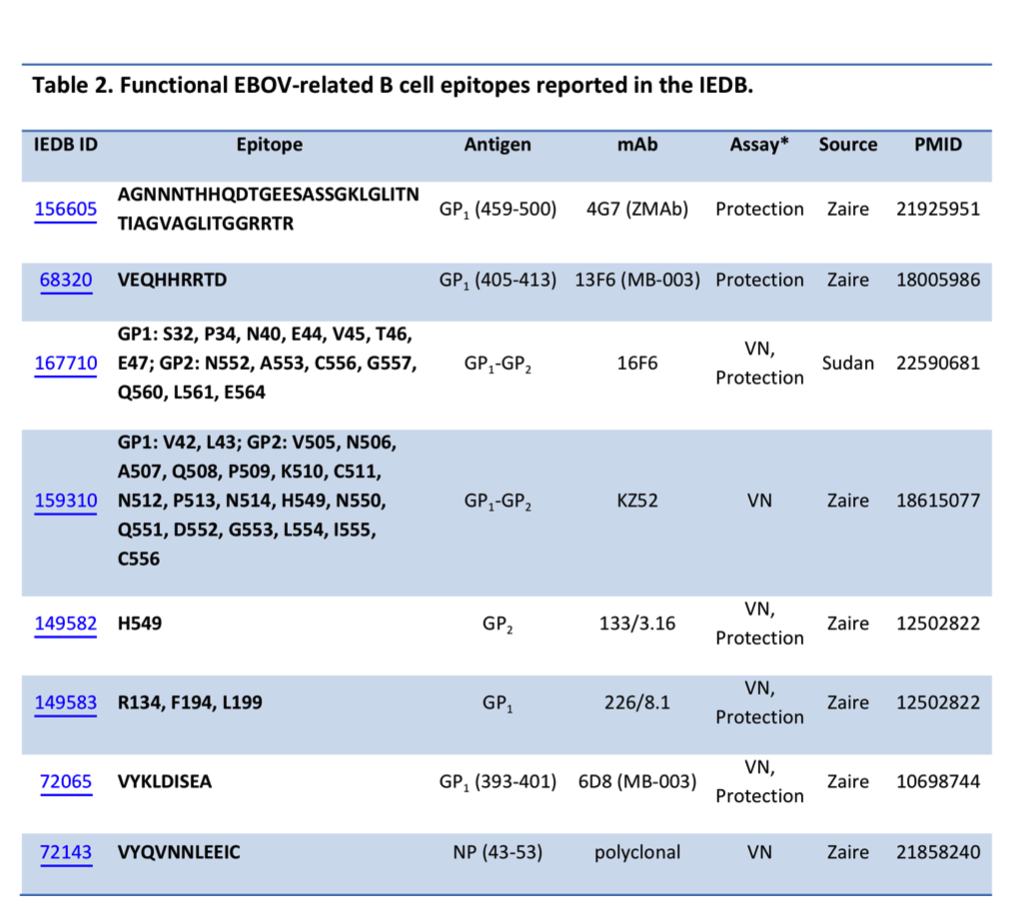
* Protection means a positive live in vivo survival or challenge assay; VN - virus neutralizing assay.
MHC binding data curated in the IEDB. Within the IEDB, epitopes are defined as distinct molecular structures reported to be positive in at least one immune-based experimental assay; therefore distinct, as we all overlapping structures are recorded as unique epitopes. The majority of EBOV peptides reported thus far have been defined in the context of MHC binding assays (1,101 positive peptides).
Indeed, 92% of all EBOV-specific data represent MHC binding assays. Table 3 provides a summary of all human alleles tested as positive to date by protein. Overall coverage is fairly broad with 38 different human class I alleles tested (including HLA-A, HLA-B and HLA-C), and data for all EBOV proteins. No human class II alleles have been reported to date.
While MHC binding does not define a T cell response (binding is requisite, but not solely sufficient to define immunogenicity), in the absence of a significant amount of T cell response data, MHC binding data, both experimental and predicted, might address an existing knowledge gap, and we therefore report on MHC class II prediction for the Ebola GP protein and plan reporting on MHC class I prediction and other Ebola proteins as well.

REFERENCES
· Baize S, Pannetier D, Oestereich L, Rieger T, Koivogui L, Magassouba N, Soropogui B, Sow MS, Keïta S, De Clerck H, Tiffany A, Dominguez G, Loua M, Traoré A, Kolié M, Malano ER, Heleze E, Bocquin A, Mély S, Raoul H, Caro V, Cadar D, Gabriel M, Pahlmann M, Tappe D, Schmidt-Chanasit J, Impouma B, Diallo AK, Formenty P, Van Herp M, Günther S. Emergence of Zaire Ebola Virus Disease in Guinea - Preliminary Report. N Engl J Med. 2014 Apr 16. [Epub ahead of print]
____________________________________________________________________________________
The report was prepared by Kerrie Vaughan, Julia Ponomarenko, and Alessandro Sette.
Comments
0 comments
Article is closed for comments.