The IEDB captures the MHC restriction of an epitope when it is either stated by the authors or demonstrated in the manuscript being curated. In addition, the context in which an experiment is performed is also described. For example, if the effector T cells are CD8+, this context will be described as MHC Class I. The breadth of MHC restriction data contained in the IEDB spans the 14 species currently having published epitope related data and, among other species, including humans, primates, rodents, cattle, horses, pigs, and chickens.
When performing a search, it is possible to easily narrow the results to include only those relevant to MHC Class I, MHC Class II, or Non Classical MHC on the Home Page. In addition, on the Results pages, a free text box allows one to enter any MHC allele name. Autocomplete functionality will attempt to match the entered text with MHC alleles having data present in the IEDB. Due to the complex nature of MHC nomenclature, we also provide the Allele finder which allows one to retrieve data only for a given serotype, haplotype, or locus, or to further specify an individual MHC allele, using standardized nomenclature.
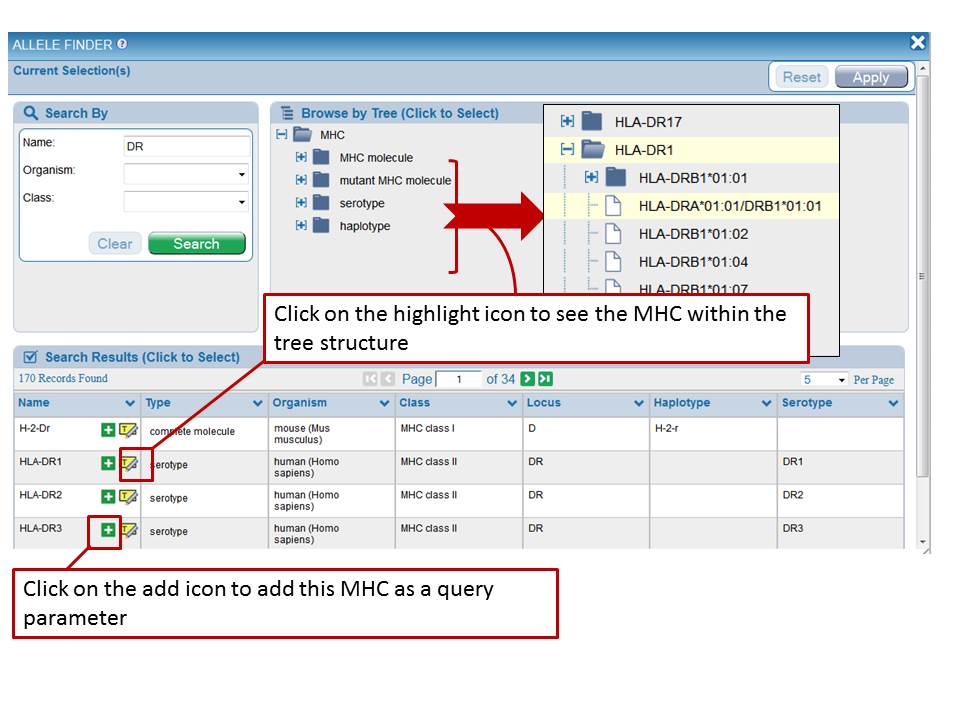
The MHC Allele Finder facilitates the selection of one or more MHC alleles.The Allele Finder presents MHC molecules in a hierarchical tree format. The Allele Finder can be found on the results page and also the Specialized Search pages. The Allele Finder may be browsed by clicking open any node of the tree or may be searched by utilizing drop down menus to select from the organisms having MHC data or by Class. One can also type a name or synonym for a specific MHC molecule, serotype, or haplotype. Browsing the tree allows users to select parent nodes of sets of MHC molecules sharing the same loci, serotype, or haplotype. As shown below, once a search has been performed, such as for "DR", the Allele Finder returns those alleles with "DR" in their name. In this case, 170 records were returned. Icons present in the Search Results allow one to select any allele by clicking the "plus sign" or to view any allele in the context of the hierarchical tree by clicking on the "Highlight" icon. Users may also click on any node of the tree to select that node as a search criteria. Once all desired alleles have been selected, users must click "Apply" to add these search parameters to their larger query of the IEDB data. Clicking "Apply" will close the Allele Finder and return the user back to the larger search interface, consisting of many other fields and Finders.
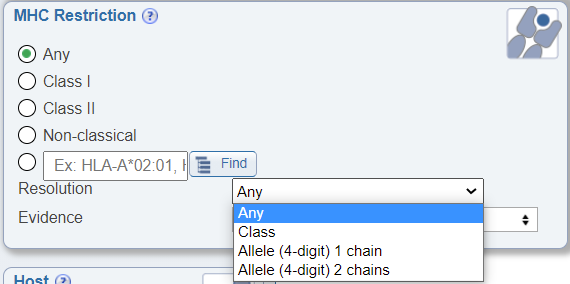
Search by MHC resolution
In the MHC Restriction search pane, a newly added drop down menu allows users to search by the resolution of the known MHC restriction. The options are Class, Allele (4 digit) 1 chain, and Allele (4 digit) 2 chains. If Class is selected, all levels of restriction will be returned in the results, including assays where the MHC restriction of an epitope was only determined to the class level; for example, 'HLA Class I' or 'BoLA Class II'. If 'Allele (4 digit) 1 chain' is selected, the returned data will include assays where the MHC restriction of an epitope was determined for one or both MHC chains to the level of 4 digits, which means that in addition to class (Class I) or loci (HLA-A) information, the precise restriction was determined to the level of 4 digits (HLA-A*02:01). For Class II alleles, this dataset will include cases where the MHC restriction was determined for the alpha or beta or both chains, such as cases where the restriction was determined to be HLA-DQA1*05:05. If 'Allele (4 digit) 2 chains' is selected, the dataset will include all MHC Class I restricted epitope data determined to the level of 4 digits and only Class II restricted epitope data where both alpha and beta paired chains were identified to the level of 4 digits, such as HLA-DQA1*05:05/DQB1*02:02. Note that all data in the IEDB is either curated from the literature or directly submitted. Thus, the MHC restriction information in each assay is provided by the authors of that data. MHC restriction may be blank or provided at the Class level, Locus level, 4 digit single chain or 2 chain levels. The same epitope may be stated by one publication to be 'HLA Class I' restricted, to be 'HLA-A restricted in another publication' and stated to be 'HLA-A*02:01' restriction in yet another publication.
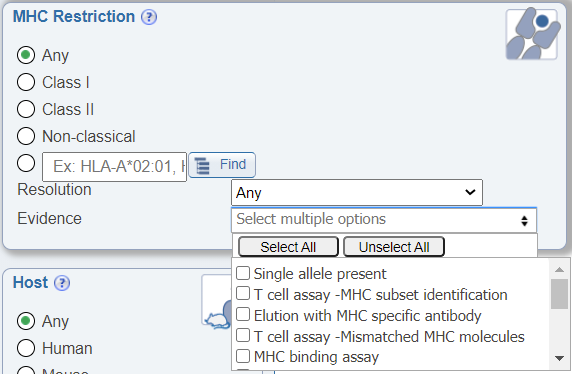
Search by MHC Evidence
In the MHC Restriction search pane, another newly added drop down menu allows users to search by the evidence of the MHC restriction information being provided. Different publications may use a variety of means to determine the MHC restriction of an epitope and we found that users wanted to be able to narrow their data accordingly. The search options include the following:
- Single allele present: The restriction of the epitope was determined based on the use of antigen presenting cells expressing a single MHC molecule or through the use of multimer technology.
- T cell assay - MHC subset identification: The restriction of the epitope was determined based on the use of antibodies to block specific MHC during a T cell assay.
- Allele/Locus-specific antibody: The restriction of the epitope was determined by the use of antibodies that recognize specific MHC groups during an MHC ligand elution assay.
- T cell assay - Mismatched MHC molecules: The restriction of the epitope was determined based on the use of different antigen presenting cells that express different MHC during a T cell assay and noting the differing effector responses.
- MHC binding assay: The restriction of the epitope was determined by either a MHC binding assay to that MHC molecule and/or being eluted from that MHC molecule.
- MHC binding prediction: The restriction of the epitope was determined by the use of binding predictions without further in vitro or in vivo experiments to confirm the restriction.
- Inferred by motif or alleles present: The restriction of the epitope was determined based on the MHC expressed on the antigen presenting cells among the responding donors.
- T cell subset identification: The restriction of the epitope was determined by the class of the T cells in the assay. For example, if CD4+ T cells responded to the epitope, the restriction was concluded as Class II. If CD8+ T cells responded to the epitope, the restriction was concluded as Class I. Note that this type of evidence only provides information on the class of MHC restriction (class I or II).
- T cell assay - Biological process measured: The restriction of the epitope was determined by the type of assay performed. For example, if proliferation was measured, the restriction was concluded as Class II. Note that this type of evidence only provides information on the class of MHC restriction (class I or II).
- Cited reference: The restriction of the epitope was previously established in a separate publication.
- Not determined: How the restriction was determined is unknown. Generally, this means the data was added to the IEDB before evidence codes were established.
Comments
0 comments
Article is closed for comments.