The Epitope Source is the natural source from which an epitope is derived. Epitope Source may be searched upon by the organism name or by the antigen name. These can be searched on the home page:
Both fields provide auto-complete functionality when one begins typing in the search boxes. The IEDB contains epitope data related to infectious diseases, allergy, autoimmunity, and transplant antigens. Therefore, the organisms from which epitopes are derived may be infectious agents, such as viruses, bacteria, and fungi, or allergens such as trees and cats, or self-organisms such as humans or mice. The antigens derived from these organisms include proteins such as haemagglutinin from influenza or Fel d1 from cat, and additionally, non-peptidic structures such as LPS from bacterial cell walls.
If the free text search does not display the organism or antigen of interest, the Molecule Finder can be used to navigate the entirety of antigens having data in the IEDB, by clicking on the "Find" icon
as found on the Search Results page of the IEDB:
Finders are available to help facilitate selections and control vocabulary usage, thus improving result outputs. At times the potential list of selections can be quite extensive, and the finders help users make selections from large lists. Multiple selections can be made when utilizing finders during a query.
The Non-Peptidic Molecule Finder is used to facilitate the selection of source antigens, immunogens, and epitopes. Records in the Source Organism Finder that is contained within the Molecule Finder come from GenPept, ChEBI, UniProt, and IEDB curators.
The Non-Peptidic Molecule Finder is designed to include two parallel trees, one for non-peptidic structures by chemical entity and the other by role. An example is shown below.
The development team determined that the most logical way to group the proteins was by organism. In order to accomplish this, the NCBI species was determined for each of the proteins in the database. For viruses and bacteria, this involved traversing the NCBI taxonomy from the sub-species (strain) level up to the species level. For each species, a set of reference proteins was selected from UniProt based upon the availability of a complete reference proteome for the species. All GenBank entries used as protein sources for epitopes in the IEDB were BLAST matched against the reference proteome set to determine their homologs. These data were used to build the protein tree in a way that mirrors a pruned version of the NCBI taxonomy. The result is a coherent tree that is divided along major taxonomic categories and is quickly traversed with proteins grouped logically below each species. The user can perform a free text search for Name and can specify the source species with the Organism Finder. The figure below shows the results for all Influenza A haemagglutinin (HA) proteins. The user can click the green plus sign  to populate the Current Selection box with their desired molecule, or they can click Highlight in Tree icon
to populate the Current Selection box with their desired molecule, or they can click Highlight in Tree icon  to see where it appears in the Protein tree, as shown below.
to see where it appears in the Protein tree, as shown below.
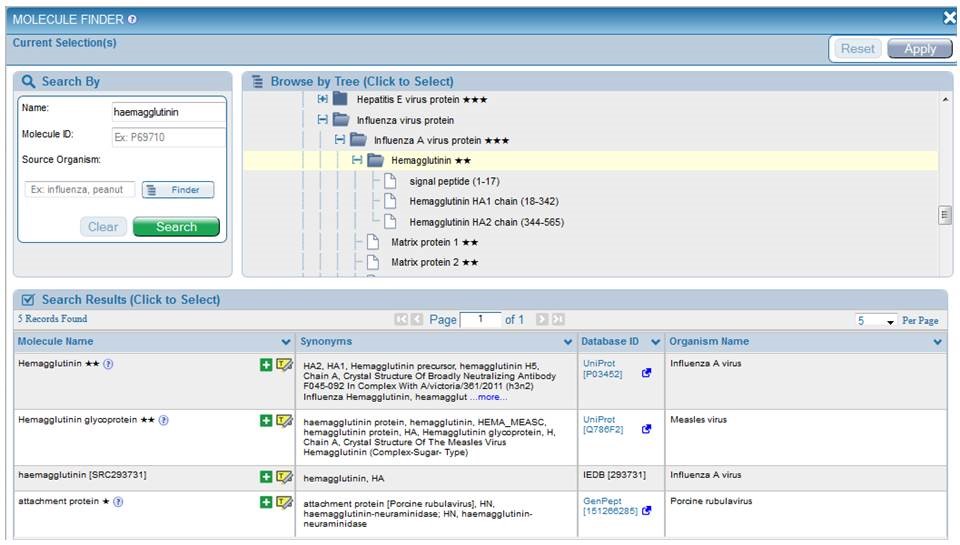
In addition to presenting proteins according to a reference proteome, proteins may be broken into processed fragments, utilizing UniProt data. Thus, Influenza A virus Hemagglutinin is presented as a signal peptide and also as HA1 and HA2. This allows users to select all epitopes from Hemagglutinin or only those epitopes from HA1, for example. The amino acid span identified by UniProt for each region is displayed next to the fragment name. For example, HA1 consists of amino acids 18-342 of the full length Hemagglutinin and is displayed as "HA1 chain (18-342)." This additional feature is especially useful for polyproteins, as are found with dengue viruses and hepatitis viruses. Please note that when searching on a processed fragment, the name displayed on the Epitopes and Antigens results tabs as the "Antigen" will remain the full protein name, such as "Hemagglutinin," but the data will reflect only epitopes present within the region the user specified, such as the HA1 chain.
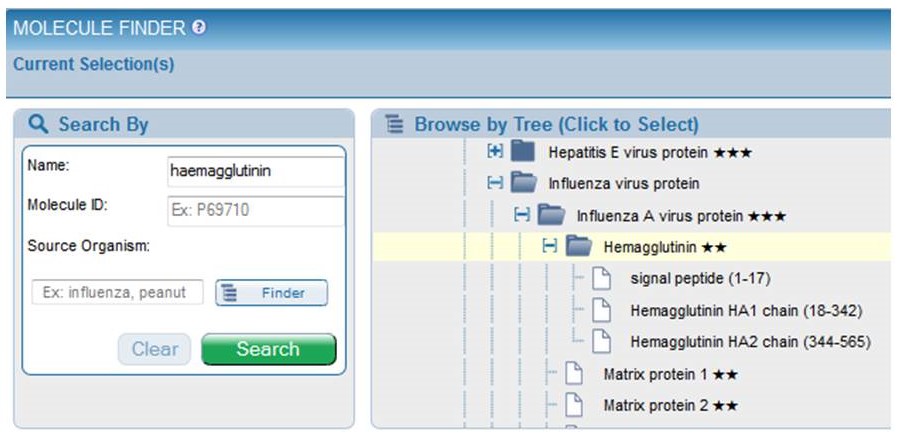
Note that the stars have been removed, as they are no longer used by UniProt.
Search by related structures
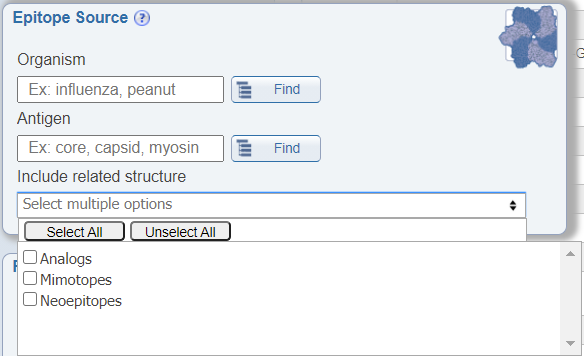
In the Epitope Source search pane, a newly added feature allows users to include structures related to their search parameters in the results that are returned. The options included are analogs, mimotopes, and neoepitopes. By checking these options, the returned results will include additional epitopes that do not specifically meet the specified search criteria, but that are related to it. For example, if one queries for Influenza A epitopes and also checks the box for 'Include related structure: analog', the epitopes that are returned will include both natural epitopes derived from Influenza A virus and artificial epitopes that are not derived from Influenza A virus, but that are analogs of Influenza A virus epitopes.
Comments
0 comments
Article is closed for comments.