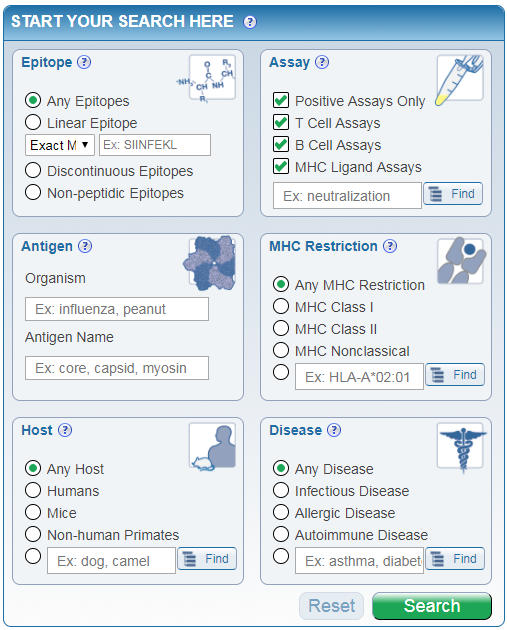
The home page search feature available in the center of the home page (Figure 1) is designed to simplify the search process for many commonly asked queries by immunologists. It is the most frequently used query mode. The approach it takes is analogous to many travel and shopping websites. IEDB users can start their search by specifying a number of fundamental and common search parameters on the home page. On the subsequent results page, users can then apply additional and more specific filters. The epitope structure choices include linear epitopes, discontinuous epitopes, or non-peptidic epitopes. For the linear epitope search, the user can prescribe the type of search desired - an exact match to the entered sequence, a substring search (described below), or a homologous peptides at sequence at identity levels of 70%, 80%, or 90% as determined by BLAST. The IEDB categorizes all assays as T cell, B cell, or MHC ligand. The MHC ligand includes binding and elution assays. Users can also select the MHC restriction for non-B cell searches. Finally, users can select the host species and the disease.
The results of the query are displayed in four tabs – Epitopes, Antigens, Assays, and References. The Epitopes tab lists the epitope description, such as the amino acid sequence for linear epitopes, its associated antigen and source organism, the number of references in which it appears, and the number of assays in which it is used. The Antigen tab lists the antigen, its source organism, and the number of related epitopes, assays, and references. The Assays tab contains three tabs, one each for T cell, B cell, and MHC ligand assays. All tabs list the IEDB-specific assay identifier, the reference in which it appears, and the related epitope. The other fields listed are those relevant for each assay type. For example, MHC restriction does not appear for B cell assays. The References tab lists the IEDB-specific reference identifier, the authors, title, abstract, and year of publication or submission. In all cases, the user can click on the IEDB-specific identifier to drill down to get additional information about the epitope, assay, or reference.
|
Figure 1. Home page Simple Search |
Once the initial results are generated, the user can filter the results using additional filtering criteria. Auto-complete fields and associated hierarchical tree views reveal the available parameter choices. In this way, users can narrow their query to specific non-peptidic epitopes, assay types, MHC alleles, hosts, or diseases. Users can also specify article type (journal article and/or submission), the author, title, PubMed ID, and year. All filters that are applied are displayed at the top of the page. Results on most pages can be downloaded as comma separated value (CSV) files that can be read using a text editor or spreadsheet application. Users also have the ability to map positive and negative results to reference proteomes as a means of visualizing immunogenic regions (see help topic on Immunome Browser).
An autocomplete feature is available for many of the fields on the lefthand side of the results page that contain additional filter criteria. This includes the non-peptidic epitope field in the Epitope box, the organism and antigen name fields in the Antigen box, the T cell, B cell, and MHC ligand assay fields in the Assay box, the specific MHC restriction field in the MHC Restriction box, the specific host field in the Host box, the specific disease field in the Disease box, and the author, title, and data fields in the Reference box. For example, as one types “hep” in the organism field in the Antigen box, several choices start to appear in a list below the text field, including “hepatitis C virus”. The user can click on a selection from the list or can continue typing to further narrow down choices. Likewise, a user may type “human” or “homo” to select “homo sapiens” in the Organism field. The list of matches includes scientific names and synonyms. These actions enable users to quickly specify their search parameters without using the finders. The autocomplete applies wherever the input field has example input in light grey. Use of the autocomplete circumvents the need to use the finders.
If the user decides not to use the autocomplete feature for a search field, they will need to use the finders. The epitope source can be prescribed for the source organism and source antigen by using the organism finder (see separate help topic) and molecule finder see separate help topic), respectively. The user can decide whether to include B cell responses, T cell responses, and/or MHC ligand results in the search (at least one must be checked). The host organism, the MHC restriction, and the MHC class can also be specified with the help of the organism finder and the allele finder (see separate help topic). The fields using finders will allow multiple selections as search criteria. In these cases the selections are treated as a set. Records will be considered a match if they include at least one of the selected values in the set. The search is executed by selecting the Search button and query results can be viewed on the updated results page described in the help topic "Understanding Search Results."
In addition to finding peptides in the database that contain the specified amino acid sequence, the substring search also finds epitopes within the input sequence itself. For example, when the sequence AELLVALENQHTIDL is submitted for a substring search, the query results, shown in Figure 2, yields five peptide sequences. Three of them (the second, third, and fifth) contain the input sequence. The two others (the first and fourth) are substrings of the input sequence that are also epitopes contained in the IEDB.
|
Figure 2. Query results for a substring search performed on the input sequence AELLVALENQHTIDL |
Comments
0 comments
Article is closed for comments.