Overview:
As of 2021, the IEDB has updated the results page to include new specialized Filter Options, which provide a set of new, or previously hidden, filter parameters for specific search purposes. These filter options are accessed at the top of the left search panel on the results page, and they give users access to additional search parameters that were previously only available in the Specialized Searches. Given that the Specialized Searches were hidden within a rarely used drop down menu, these have been simultaneously retired, and the Filter Options now contain additional search parameters to help users further refine their queries for T cell, B cell and MHC data. Additionally, new search features have been added based on user feedback.
How do the 'Filter Options' work?
There are 4 different filter options, specific to user needs; Default, T Cell, B Cell and MHC. The filter is selected based on the original query from the IEDB homepage - for example, if one selects 'T cell assays', the T Cell option will be automatically selected. If no parameters on the homepage were changed, the 'Default' view will be presented on the results page, which offers general Filter Options in the left panel, as described below. If a new view is selected, which is tailored to either T cell, B cell, or MHC data, the search pane color will be updated to show the user what search panes have changed from the 'Default' view.

Reminder:
Remember when changing filters, you will need to select the green 'Search' button to ensure that your new filters are applied to your search.
What is New?
Each Filter Option is tailored to its named type to include both previously hidden and newly available search parameters, as described in detail below.
Newly added search parameters for all Filter Options
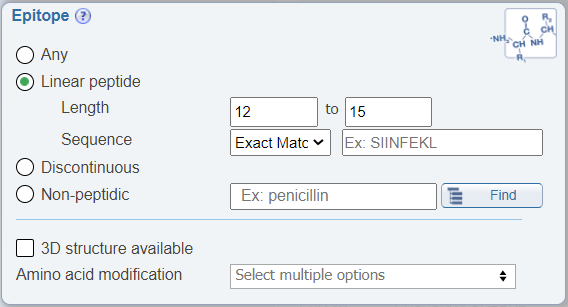
Search by peptide length
In the Epitope search pane, users can now limit their search to a range of epitope lengths.
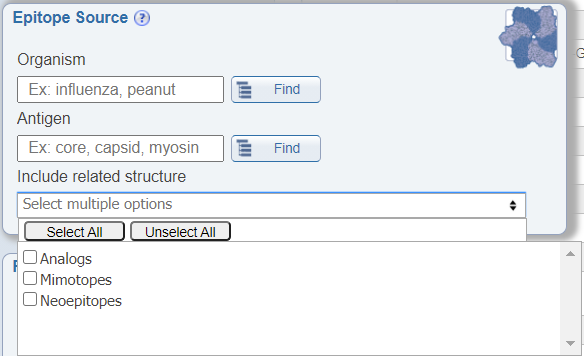
Search by related structures
In the Epitope Source search pane, a newly added feature allows users to include structures related to their search parameters in the results that are returned. The options included are analogs, mimotopes, and neoepitopes. By checking these options, the returned results will include additional epitopes that do not specifically meet the specified search criteria, but that are related to it. For example, if one queries for Influenza A epitopes and also checks the box for 'Include related structure: analog', the epitopes that are returned will include both natural epitopes derived from Influenza A virus and artificial epitopes that are not derived from Influenza A virus, but that are analogs of Influenza A virus epitopes.
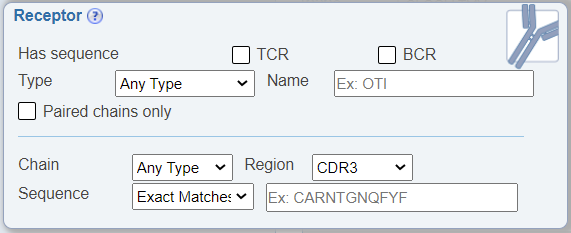
Search by receptor chains
In the Receptor Search pane, a newly added feature allows one to search for BCR or TCR sequences that are paired, which means that both heavy and light BCR or alpha and beta TCR chains were sequenced. This is accomplished by checking the new check box labeled as 'Paired chains only'. The default is for this option to be unchecked, which will return all receptor sequence data, whether both chains were sequenced or not.
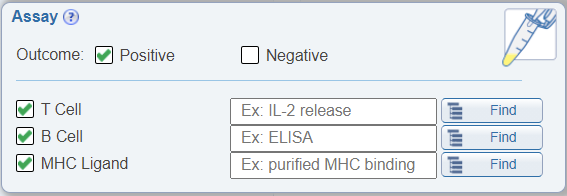
Search for negative data
In the Assay search pane, it is now possible to search for negative data. Previously, one was able to search for all data or positive data. There are now two check boxes in the Assay search pane; one for Positive and one for Negative data. The default is to have Positive checked.
Search by MHC resolution
In the MHC Restriction search pane, a newly added drop down menu allows users to search by the resolution of the known MHC restriction. The options are Class, Allele (4 digit) 1 chain, and Allele (4 digit) 2 chains. If Class is selected, all levels of restriction will be returned in the results, including assays where the MHC restriction of an epitope was only determined to the class level; for example, 'HLA Class I' or 'BoLA Class II'. If 'Allele (4 digit) 1 chain' is selected, the returned data will include assays where the MHC restriction of an epitope was determined for one or both MHC chains to the level of 4 digits, which means that in addition to class (Class I) or loci (HLA-A) information, the precise restriction was determined to the level of 4 digits (HLA-A*02:01). For Class II alleles, this dataset will include cases where the MHC restriction was determined for the alpha or beta or both chains, such as cases where the restriction was determined to be HLA-DQA1*05:05. If 'Allele (4 digit) 2 chains' is selected, the dataset will include all MHC Class I restricted epitope data determined to the level of 4 digits and only Class II restricted epitope data where both alpha and beta paired chains were identified to the level of 4 digits, such as HLA-DQA1*05:05/DQB1*02:02. Note that all data in the IEDB is either curated from the literature or directly submitted. Thus, the MHC restriction information in each assay is provided by the authors of that data. MHC restriction may be blank or provided at the Class level, Locus level, 4 digit single chain or 2 chain levels. The same epitope may be stated by one publication to be 'HLA Class I' restricted, to be 'HLA-A restricted in another publication' and stated to be 'HLA-A*02:01' restriction in yet another publication.
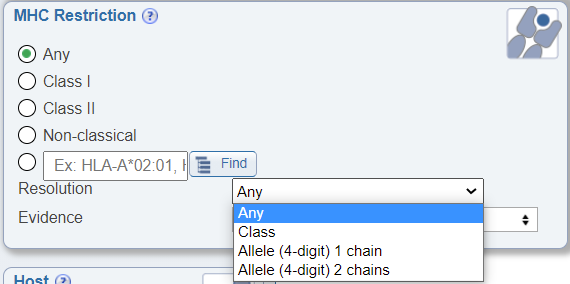
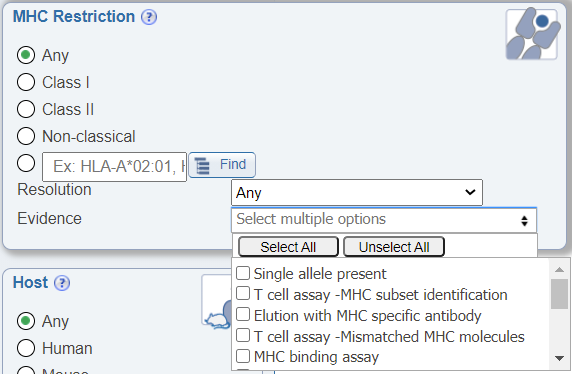
Search by MHC Evidence
In the MHC Restriction search pane, another newly added drop down menu allows users to search by the evidence of the MHC restriction information being provided. Different publications may use a variety of means to determine the MHC restriction of an epitope and we found that users wanted to be able to narrow their data accordingly. The search options include the following:
- Single allele present: The restriction of the epitope was determined based on the use of antigen presenting cells expressing a single MHC molecule or through the use of multimer technology.
- T cell assay - MHC subset identification: The restriction of the epitope was determined based on the use of antibodies to block specific MHC during a T cell assay.
- Allele/Locus-specific antibody: The restriction of the epitope was determined by the use of antibodies that recognize specific MHC groups during an MHC ligand elution assay.
- T cell assay - Mismatched MHC molecules: The restriction of the epitope was determined based on the use of different antigen presenting cells that express different MHC during a T cell assay and noting the differing effector responses.
- MHC binding assay: The restriction of the epitope was determined by either a MHC binding assay to that MHC molecule and/or being eluted from that MHC molecule.
- MHC binding prediction: The restriction of the epitope was determined by the use of binding predictions without further in vitro or in vivo experiments to confirm the restriction.
- Inferred by motif or alleles present: The restriction of the epitope was determined based on the MHC expressed on the antigen presenting cells among the responding donors.
- T cell subset identification: The restriction of the epitope was determined by the class of the T cells in the assay. For example, if CD4+ T cells responded to the epitope, the restriction was concluded as Class II. If CD8+ T cells responded to the epitope, the restriction was concluded as Class I. Note that this type of evidence only provides information on the class of MHC restriction (class I or II).
- T cell assay - Biological process measured: The restriction of the epitope was determined by the type of assay performed. For example, if proliferation was measured, the restriction was concluded as Class II. Note that this type of evidence only provides information on the class of MHC restriction (class I or II).
- Cited reference: The restriction of the epitope was previously established in a separate publication.
- Not determined: How the restriction was determined is unknown. Generally, this means the data was added to the IEDB before evidence codes were established.
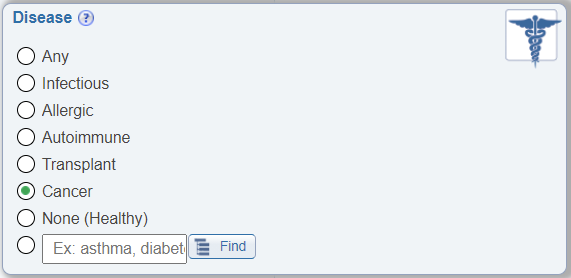
Search by Cancer
In the Disease search pane, a new radio button to allow easy search for epitopes tested for immune recognition by hosts with cancer has been added.
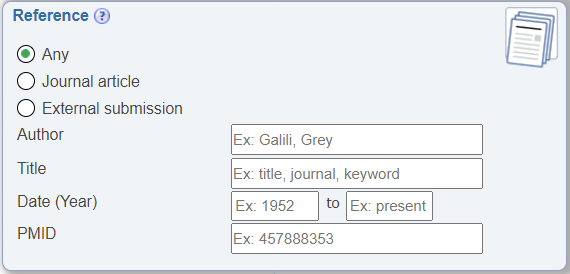
Search by PMID
In the Reference search pane, the ability to search by PMID was made more prominent by having it displayed by default. Previously, it was hidden until users selected the 'Journal article' radio button.
T Cell Specific Filter Options
In addition to the above new search features, the T Cell view also renames the 'Receptor' search pane to be 'TCR' with TCR-specific drop down menus that no longer include BCR-related search parameters, for example chain type search options are now alpha and beta. The 'Assay' search pane has been renamed as 'T Cell Assay' and displays radio buttons for commonly searched T cell assay types, which were previously found within the Assay Finder. Additionally, a new check box for 'direct ex vivo detection' was added that allows one to search for T cell assay data where effector cells were tested directly ex vivo without further in vitro restimulation.
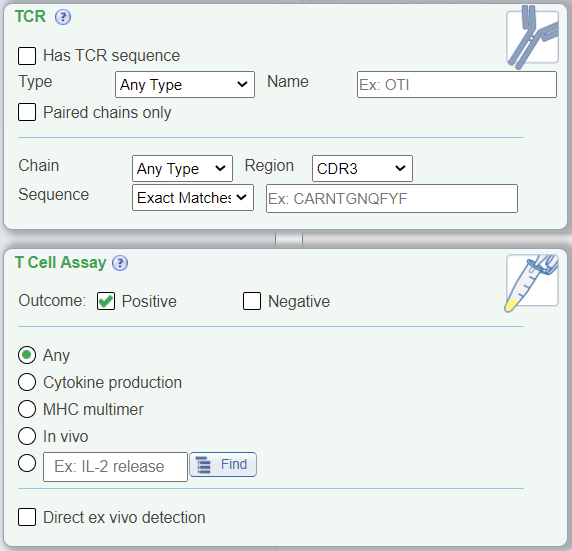
B Cell Specific Filter Options
In addition to the above new search features, the B Cell view also renames the 'Receptor' search pane to be 'Antibody/BCR' with BCR specific drop down menus that no longer include TCR related search parameters, for example chain type search options are now heavy and light. The 'Assay' search pane has been renamed as 'B Cell Assay' and displays radio buttons for commonly searched B cell assay types, which were previously found within the Assay Finder. Additionally, a new drop down menu called 'Antibody isotype' has been added that allows users to search by the isotype of the antibody response that was measured. Lastly, the B Cell Filter does not display the 'MHC Restriction' search pane, as it is not relevant to this search type.
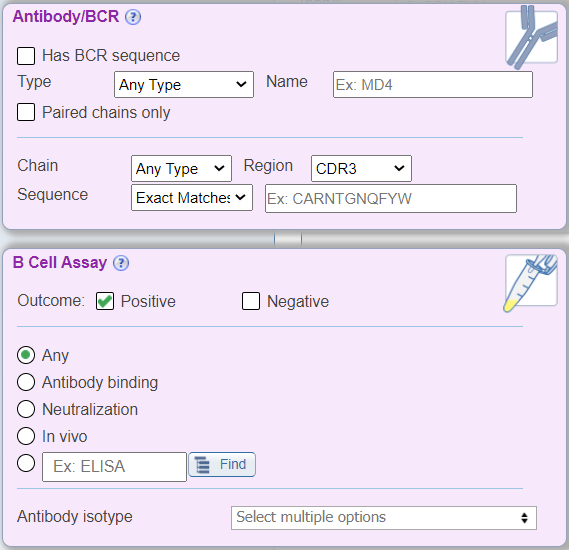
MHC Specific Cell Filter Options
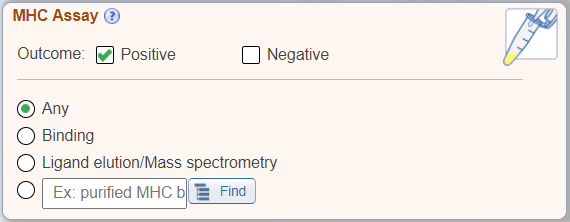
In addition to the above new search features, the 'Assay' search pane has been renamed as 'MHC Assay Type' and displays radio buttons for commonly searched MHC specific assay types, which were previously found within the Assay Finder.
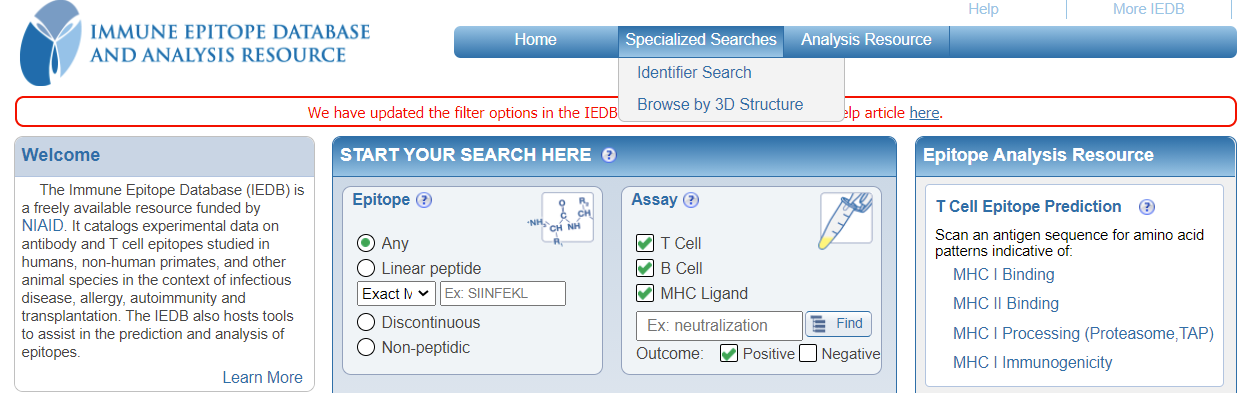
What happened to the 'Specialized Searches'?
The 'Specialized Searches' menu has been reviewed extensively and the most commonly used search features have been incorporated into the new filter options, as described above. The following specialized search options have been removed from the dropdown menu:
- Epitope Details
- T Cell Assay Details
- B Cell Assay Details
- MHC Assay Details
You can continue to use the below options from the 'Specialized Searches' dropdown menu:
- Identifier Search
- Browse by 3D Structure
It is also possible to fully export all of the data in the IEDB in the Database Export page: https://www.iedb.org/database_export_v3.php
Comments
0 comments
Please sign in to leave a comment.